Rosetta store: Nested like in R
keywords jamovi, R, Mixed Models, Multilevel Models, Nested design, Nested levels
GAMLj version ≥ 1.0.0
Issue
I have nested clustering variables (nested levels) and I am not sure the model is considering the nesting correctly.
Short answer
Be sure that every cluster, at each level, has a unique value (factor level). If so, the model considers the nesting correctly
Long answer
Introduction
Mixed models are often used to analyze multilevel designs, in which
several clustering variables appear. When more than one clustering
variable exists, clusters can be nested or cross-classified. See
Mixed Models: Subjects by Stimuli random
effects for some examples. In case of nested models, users sometimes
wonder how to signal to GAMLj that the
clustering variables are nested, meaning that they define different
levels of a multilevel design. This uncertainty is often supported by
the fact that lmer() command of R package lme4
seems to require specifying the nested nature of the clustering
variables with special notation (see below). However, this is not the
case. Nested clustering can be specified both in lmer()
command and in GAMLj without any special
notation (model formula notation), and the models are correctly
estimated taking into the account the nested nature of the design. Let’s
see a toy example to clarifying the issue.
A quick three-level example
Assume you have three schools, within each school three classes, and
pupils within classes. Schools are coded in variable school
as A, B, and C. Assume you coded,
within each school, the three classes as 1, 2,
and 3. Your dataset will look like this.
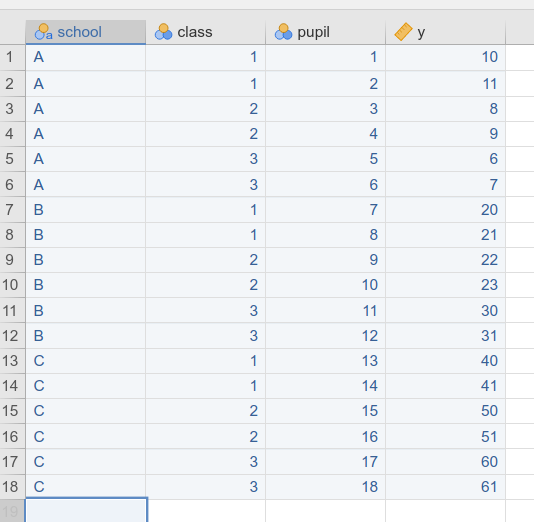
This means that the design features 3 schools, and 9 classes.
Usually, you want to estimate the random coefficients (in this simple
case only the intercepts) across schools and classes. If you run a model
in GAMLj using these data, however, the
model will pool together all pupils in class 1, all pupils
in class 2 an so on, irrespective of their school, as if
pupils of different schools belonged to the same class. Indeed, in GAMLj you would have 3 schools and 3 classes
:
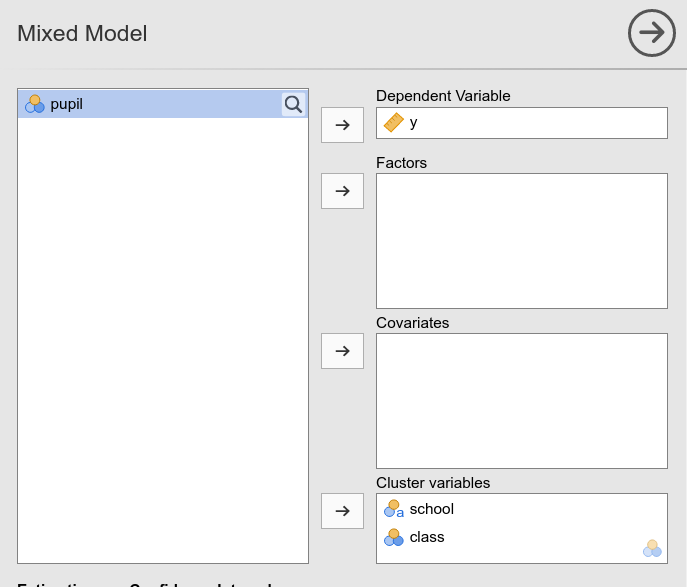

This is equivalent to run in R lme4 command
library(lmerTest)
data$school<-factor(data$school)
data$class<-factor(data$class)
mod<-lmer(y~1+(1|school)+(1|class),data=data)
print(mod,digits=5)## Linear mixed model fit by REML ['lmerModLmerTest']
## Formula: y ~ 1 + (1 | school) + (1 | class)
## Data: data
## REML criterion at convergence: 117
## Random effects:
## Groups Name Std.Dev.
## school (Intercept) 21.1056
## class (Intercept) 3.9018
## Residual 4.8291
## Number of obs: 18, groups: school, 3; class, 3
## Fixed Effects:
## (Intercept)
## 27.833Solution
To avoid this issue, R lmer() allows using a different
notation to specify the nesting. Namely
mod<-lmer(y~1+(1|school/class),data=data)
print(mod,digits=5)## Linear mixed model fit by REML ['lmerModLmerTest']
## Formula: y ~ 1 + (1 | school/class)
## Data: data
## REML criterion at convergence: 87.6
## Random effects:
## Groups Name Std.Dev.
## class:school (Intercept) 6.6144
## school (Intercept) 20.8497
## Residual 0.7071
## Number of obs: 18, groups: class:school, 9; school, 3
## Fixed Effects:
## (Intercept)
## 27.833As you can see, the model has now 9 classes and 3 schools, as
intended. However, the only thing that this notation does is to build a
new variable class:school which assigns unique values to
each class. Indeed, we can do that in R and in GAMLj to obtain exactly the same results.
In R:
data$unique_class<-apply(cbind(data$class,data$school),1,paste,collapse="_")
data$unique_class<-factor(data$unique_class)
mod<-lmer(y~1+(1|school)+(1|unique_class),data=data)
print(mod,digits=5)## Linear mixed model fit by REML ['lmerModLmerTest']
## Formula: y ~ 1 + (1 | school) + (1 | unique_class)
## Data: data
## REML criterion at convergence: 87.6
## Random effects:
## Groups Name Std.Dev.
## unique_class (Intercept) 6.6144
## school (Intercept) 20.8497
## Residual 0.7071
## Number of obs: 18, groups: unique_class, 9; school, 3
## Fixed Effects:
## (Intercept)
## 27.833In jamovi:

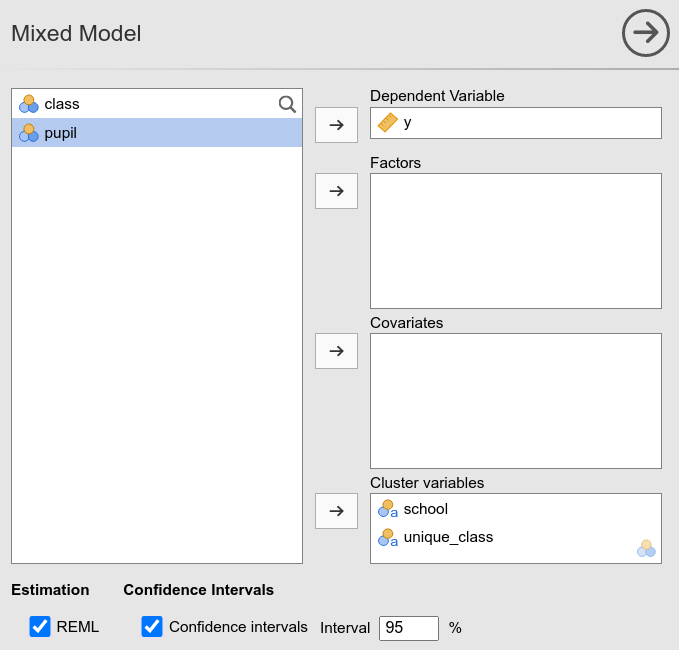
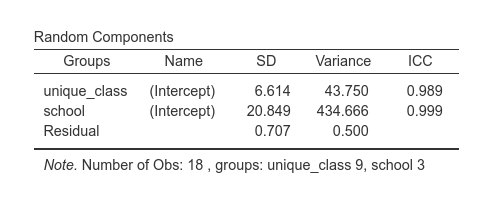
Indeed, the new variable unique_class is simply
assigning unique values to the classes.

Recap
When dealing with nested levels, be sure that each group (cluster) is coded with a unique code, and the nesting structure will be automatically taken into the account by the model.
Rosetta’s files
- Rosetta store: contrasts
- Rosetta store: Generalized Mixed
- Rosetta store: Nested like in R
- Rosetta store: moderation
Comments?
Got comments, issues or spotted a bug? Please open an issue on GAMLj at github or send me an email